Research
Selected Research Highlights

Generative AI in Healthcare
LLMs in Medical Imaging - An early investigation
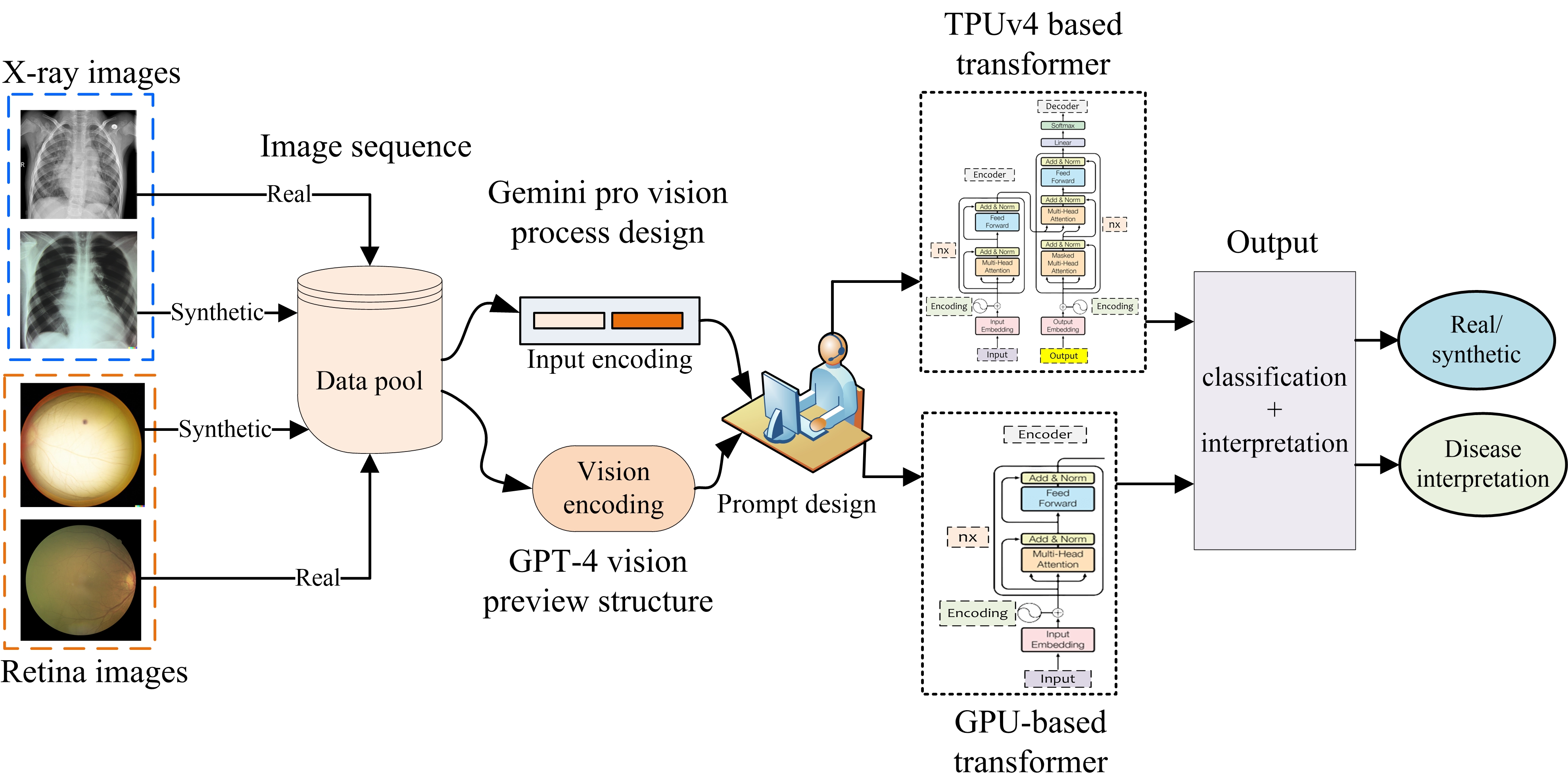
There is a temptation to use generative models to synthesize realistic-looking medical image data, while on the other hand, the ability to identify synthetic image data in a pool of data is also significantly important. In this study, we explore the potential of the Gemini (gemini-1.0-pro-visionlatest) and GPT-4V (gpt-4-vision-preview) models for medical image analysis using two modalities of medical image data. Utilizing synthetic and real imaging data, both Gemini AI and GPT-4V are first used to classify real versus synthetic images, followed by an interpretation and analysis of the input images. In this specific experiment, Gemini was able to perform slightly better than the GPT-4V on the classification task. In contrast, responses associated with GPT-4V were mostly generic in nature. Our early investigation presented in this work provides insights into the potential of MLLMs to assist with the classification and interpretation of retinal fundoscopy and lung X-ray images. We also identify key limitations associated with the early investigation study on MLLMs for specialized tasks in medical image analysis.
See relevant publications here
Generative Adversarial Networks for Medical Imaging
Medical Image Super-Resolution using GANs
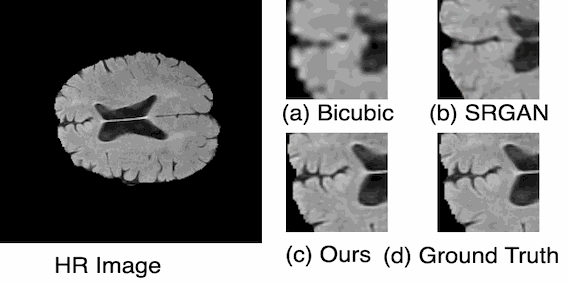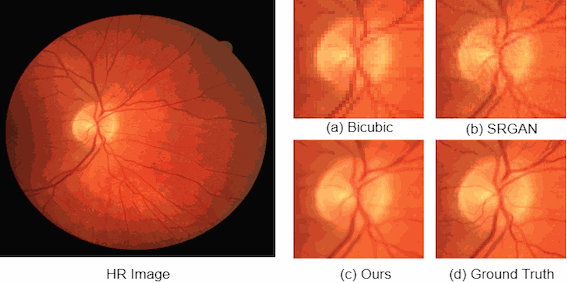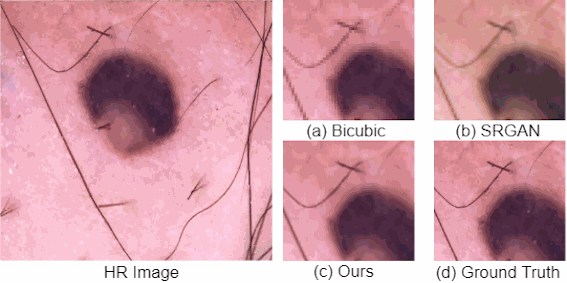
For medical image analysis, there is always an immense need for rich details in an image. In medical imaging, acquiring high-resolution images is challenging and costly as it requires sophisticated and expensive instruments, trained human resources, and often causes operation delays. Deep learning based super resolution techniques can help us to extract rich details from a low-resolution image acquired using the existing devices. We propose a new Generative Adversarial Network (GAN) based architecture for medical images, which maps low-resolution medical images to high-resolution images. The proposed architecture is divided into three steps. In the first step, we use a multi-path architecture to extract shallow features on multiple scales instead of single scale. In the second step, we use a ResNet34 architecture to extract deep features and upscale the features map by a factor of two. In the third step, we extract features of the upscaled version of the image using a residual connection-based mini-CNN and again upscale the feature map by a factor of two. The progressive upscaling overcomes the limitation for previous methods in generating true colors. Finally, we use a reconstruction convolutional layer to map back the upscaled features to a high-resolution image. Our addition of an extra loss term helps in overcoming large errors, thus, generating more realistic and smooth images. We evaluate the proposed architecture on four different medical image modalities: (1) the DRIVE and STARE datasets of retinal fundoscopy images, (2) the BraTS dataset of brain MRI, (3) the ISIC skin cancer dataset of dermoscopy images, and (4) the CAMUS dataset of cardiac ultrasound images. The proposed architecture achieves superior accuracy compared to other state-of-the-art super-resolution architectures.
Read about this work in Nature Scientific Reports
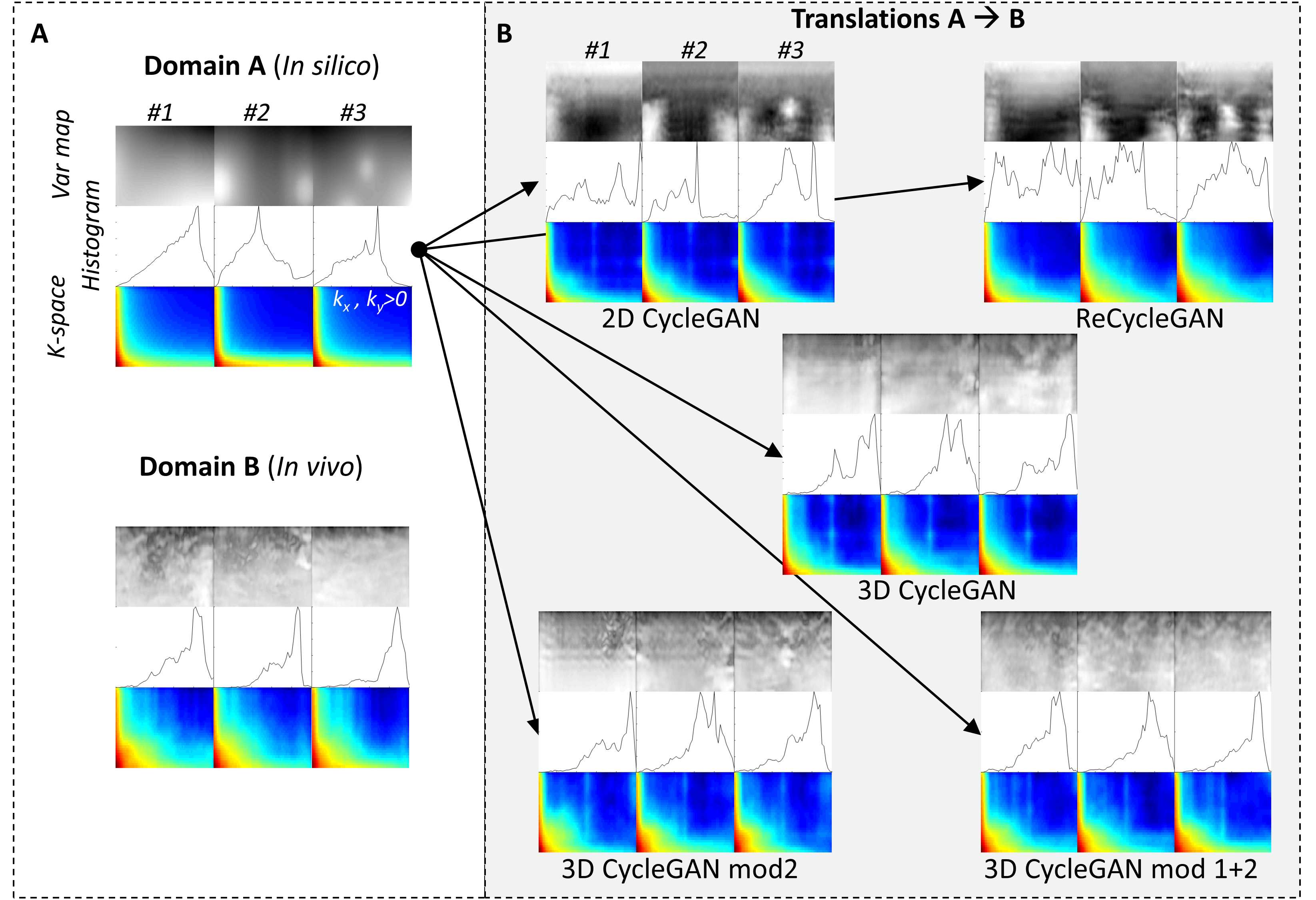
3D CycleGAN model for ultrasound image sequences of intra-muscular skeletal muscle contractions
Advances in sports medicine, rehabilitation applications and diagnostics of neuromuscular disorders are based on the analysis of skeletal muscle contractions. Medical imaging techniques have transformed the study of muscle contractions by allowing to learn the complex patterns of muscle activation. We use deep learning (3D cycleGAN) to model the authentic intra-muscular skeletal muscle contraction pattern using domain-to-domain translation between in silico (simulated) and in vivo (experimental) image sequences of skeletal muscle contraction dynamics. Our results show that there are large differences between the spatial features of in silico and in vivo data, and that a model could be trained to generate authentic spatio-temporal features similar to those obtained from in vivo experimental data.
Read about this work in Biomedical Engineering Online.
Translation of atherosclerotic disease using CycleGAN
Read more about translation of atherosclerotic disease published in Elsevier
GANs for lungs segmentation in Chest X-rays
Chest X-ray procedures are considered to be the most popular for diagnosis of chest related diseases. We as a machine learning and medical imaging community, have seen extraordinary interest in the chest x-rays anlaysis and segmentation tasks. For any diagnosis on chest x-rays, accuracte segmentation of the biological object is fundamental. Here, we show how we can use Generative Adversarial Networks (GANs) to perform segmentation of lungs within chest x-rays. The generator of the GAN generates a segmented mask of a given chest x-ray. Once the generator is trained to generate realistic looking masks, the GAN can now be used to perform segmentation of the lungs on new chest x-ray images.
The full paper is available on IEEE Access
GANs for retinal images
We propose a new Generative Adversarial Network for Medical Imaging (MI-GAN). The MI-GAN generates synthetic medical images and their segmented masks, which can then be used for the application of supervised analysis of medical images. This work presents MI-GAN for synthesis of retinal images. The MI-GAN method generates precise segmented retinal images better than the existing techniques.
Read about this work in Springer NCA
Read the book chapter on Springer
Urdu text processing (UTPro)
UHaT Data: Urdu Handwritten Text Dataset
Urdu is the national language of Pakistan and one of the major languages of the world. Research on Urdu hand-written text processing has not been addressed before primarily due to lack of datasets and lack of baseline research. This project is aimed at:
- developing a freely available dataset of Urdu handwritten characters. The dataset developed so far comprises of isolated hand-written characters of Urdu, comprising of 1000 images per character.
- Build a deep learning framework for recognition of the isolated words.
The UHaT dataset can be obtained for free from Kaggle
The UHaT dataset is now featured on the IAPR Technical Committee 11 webpage.
Acknowledgements: This works has been funded by the research grant 21-790/SRGP/R&D/HEC/2016 of Higher Education Commission Islamabad (HEC).
Read more about this work in Springer
Urdu Speech Recognition
Automatic Speech Recognition (ASR) has been a topic of interest for the last several decades. Today, ASR technology is widely used in our daily life as in Android/iOS devices as robust speech recognition frameworks are available for developed languages. However, for Urdu language, the research work is less developed. This project provides:
- A freely available speech corpus of Urdu isolated words. Currently, the corpus consists of 250 words. In more refined form, a corpus of 100 words can be obtained for free use (available from download section of this website).
- This work provides a suitable baseline approach based on extraction of Mel-frequency cepstral coefficients and then used for training speech recognition engine such as Hidden Markov Models. The results obtained thus are good baseline for further research work. Particular topic of interest is continuous speech recognition of Urdu.
The dataset can be obtained for free from Kaggle